St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
About the Klco Lab
Myeloid tumors in children are commonly caused by a different set of mutations than those seen in adults, but the genetics of pediatric myeloid tumors are not well understood. Our laboratory is focused on defining the molecular and biological causes of this class of tumors. We build collaborative partnerships and use genomics to identify new classes of mutations that lead to these cancers. Our work aims to transform the diagnosis and management of these diseases in children.

Our research summary
Uniquely positioned at the interface of basic research and clinical care, our laboratory deciphers the underlying mechanisms of pediatric myeloid tumors. We use genomic sequencing and -omics analyses to identify novel and recurrent mutations coupled with in vitro and in vivo strategies to model and define the molecular mechanisms associated with these genetic alterations. The goal of our research is to improve the diagnosis and risk stratification of myeloid tumors and identify molecular pathways that may be amenable to therapeutic targeting. Current projects leverage human model systems, CRISPR-Cas9 technology, degron systems, and in vivo murine models to decipher the mechanisms of disease development and progression. Collectively, our work aims create representative models that can be used to discover the mechanisms of disease development, progression and for the establishment of pre-clinical models of myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) to support therapeutic development.
Identification of genetic drivers in pediatric myeloid tumors
The myeloid tumors (notably myelodysplastic syndromes and acute myeloid leukemia) that develop in children commonly have a different set of genetic alterations than those seen in adults. Yet very little is known about the genetics of pediatric myeloid tumors. Our laboratory has published landmark studies on core binding factors in AMLs, pediatric AML, pediatric MDS, and pediatric therapy-related myeloid neoplasms to comprehensively define the somatic and germline alterations that drive these neoplasms.
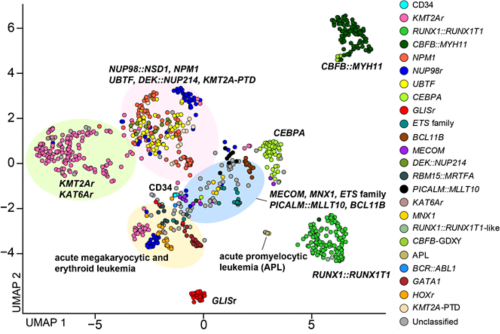
UMAP plot of the pAML cohort (n = 887) and cord blood CD34+ cells (normal controls: n = 5) using the top 315 variable genes.
SAMD9 and SAMD9L
Our studies on pediatric MDS identified a new class of predisposition genes (SAMD9 and SAMD9L) in nearly 20% of children with MDS. These children universally also have somatic loss of chromosome 7 (monosomy 7), which can further drive disease progression. Human models have demonstrated that the expression of these mutations leads to increases in inflammation that cause cell death. Murine models have demonstrated that the expression of SAMD9 mutations leads to bone marrow failure and chromosomal deletions that recapitulated clinical manifestation of SAMD9/SAMD9L pediatric MDS cases. These findings have an immediate impact on patient care, both for diagnosis and monitoring.
UBTF
We recently identified tandem duplications involving exon 13 of UBTF gene (UBTF-TD) as a recurrent and category-defining genetic alteration enriched in pediatric AML. Our screening strategies found UBTF-TD alterations present in cases of pediatric AML at diagnosis and at relapse, which led to broad recognition of UBTF-TD in adult AML and pediatric MDS. The development of human and murine systems to model UBTF-TD driven AML have deepened our mechanistic understanding of how UBTF-TD drives leukemogenesis through genomic mislocalization to HOX regions. Moreover, our model systems have led to the identification of Menin inhibitors as effective therapies in our pre-clinical models. These findings pave the way for significant advancements in combating pediatric myeloid neoplasms.
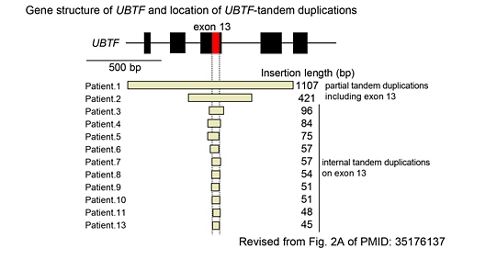
Gene structure of UBTF and location of UBTF-tandem duplications
Classification of pediatric AML
With these findings of new molecular categories that are not covered by the current WHO classification updated in 2022, we have worked on establishing a new genomic framework that comprehensively incorporates pediatric-specific driver alterations. This framework consists of 23 molecular categories defined by mutually exclusive driver alterations, covering 91% of pediatric AML. This category not only provides a detailed understanding of the biology behind each subtype but can also be leveraged to predict the prognosis with granularity. We are expanding our dataset to further characterize genetic landscape of pediatric AML associated with each molecular category.

Evolution of myeloid tumors
We strive to identify the genetic alterations that drive relapsed pediatric AML, deciphering the clonal architecture and progression of myeloid tumors from diagnosis to relapse in response to different chemotherapeutic pressures. Single cell technologies, deep sequencing and cell free DNA testing help define the spectrum of genetic mutations in pediatric tumors and how they change over time as a result of chemotherapy. This approach not only aids in our understanding of the tumor response at a granular level but facilitates the establishment of new benchmarks for minimal residual disease testing. Much of this work is in collaboration with Dr. Xiaotu Ma at St Jude.

Selected Publications
Contact us
Jeffery M. Klco, MD, PhD
Member, St. Jude Faculty
Department of Pathology
MS342, Room D4047B
St. Jude Children’s Research Hospital
Follow Us

Memphis, TN, 38105-3678 USA GET DIRECTIONS