St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Overview
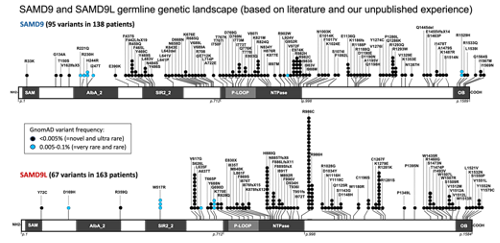
The SAMD9/SAMD9L database describes all the pathogenic germline variants and the associated clinical phenotypes reported till date. The database is an initiative to serve the medical and research community by offering a comprehensive compilation of all known information about SAMD9 and SAMD9L genes. The collection of information contains genotype, phenotype, acquired somatic mutations, karyotypic changes, gnomAD population frequency, and functional relevance about mutations known to cause SAMD9/SAMD9L associated syndromes. This website has been created in December 2019 and will be continuously updated.
SAMD9 and SAMD9L (SAMD9/9L) genes are paralogue genes on chromosome 7q involved in viral host defense, endosome formation, and cellular proliferation. Germline mutations in both genes are associated with diverse clinical phenotypes and a risk for myelodysplastic syndromes with non-random loss of chromosome 7. The full clinical picture, penetrance, and clinical outcomes of patients are not well defined. Furthermore, the physiological function of SAMD9/9L genes, as well as the mechanistic link between germline mutations and disease remain elusive.
Background
On January 27, 2024, a virtual conference for SAMD9 & SAMD9L patients/families and the medical community took place. The conference was hosted by Dr. Marcin Wlodarski and the BMF Program at St. Jude Children’s Research Hospital. The conference was separated into 3 sections: clinical education, patient advocacy and the breakout sessions. The breakout sessions were divided into 2 groups: SAMD9 and SAMD9L. More than 230 people from 27 countries registered. The registrants included roughly 113 Patients/Family Members, 68 Medical Providers, 19 Genetic Counselors, 21 Scientists, and 13 in the Other category. When asked which of the two breakout sessions registrants were more interested in attending, 77 chose SAMD9, while 67 chose SAMD9L.
On November 1 – 2, 2019, we hosted a first international meeting on SAMD9/SAMD9L disorders that brought together the international body of SAMD9/SAMD9L experts in order to assemble a network of investigators who are focused on the study of these genes as well as associated syndromes.
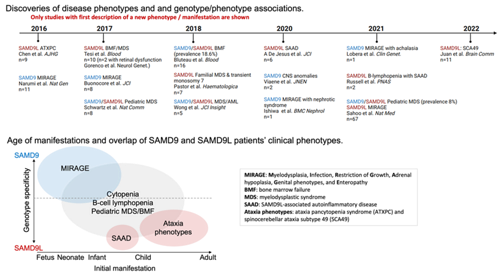
Timeline and discovery of phenotypes
Publications
Sahoo SS, Erlacher M, Wlodarski MW. Genetic and Clinical Spectrum of SAMD9 and SAMD9L Syndromes: from Variant Interpretation to Patient Management. Blood. 2024 Oct 30:blood.2022017717. doi: 10.1182/blood.2022017717. Epub ahead of print. PMID: 39475954.