St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Influenza A viruses in feral Canadian ducks: Extensive reassortment in nature
The current dogma of influenza accepts that feral aquatic birds are the reservoir for influenza A viruses. Although the genomic information of human influenza A viruses is increasing, little of this type of data is available for viruses circulating in feral waterfowl. Here, we present the genetic characterization of 35 viruses isolated from wild Canadian ducks from 1983 to 2000 as the first attempt at a comprehensive genotypic analysis of influenza viruses isolated from feral ducks. We demonstrated that influenza virus genes circulating in Canadian ducks have achieved evolutionary stasis. The majority of these duck virus genes clustered in distinct North American clades; however, some H6 and H9 genes clustered with those from Eurasian viruses. Genes appeared to reassort in a random fashion. None of the genotypes identified remained present throughout all of the years examined. We observed that most PA and PB2 genes that crossed into swine clustered in one phylogenetic grouping. Additionally, we identified matrix genes that branch very early in the evolutionary tree. These findings demonstrate the diversity of the influenza virus gene pool in Canadian ducks and suggest that genes which cluster in specific phylogenetic groupings in the PB2 and PA genes can be used for markers of viruses with the potential for crossing the species barrier. A more comprehensive study of this important reservoir is needed to provide further insight into the genomic composition of viruses that cross the species barrier, which would be a useful component to pandemic planning.
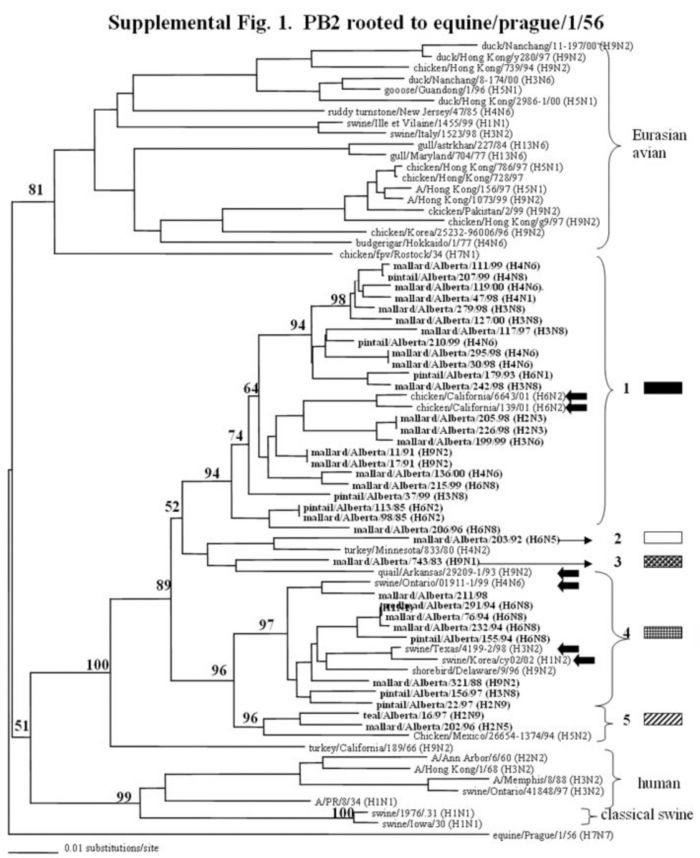
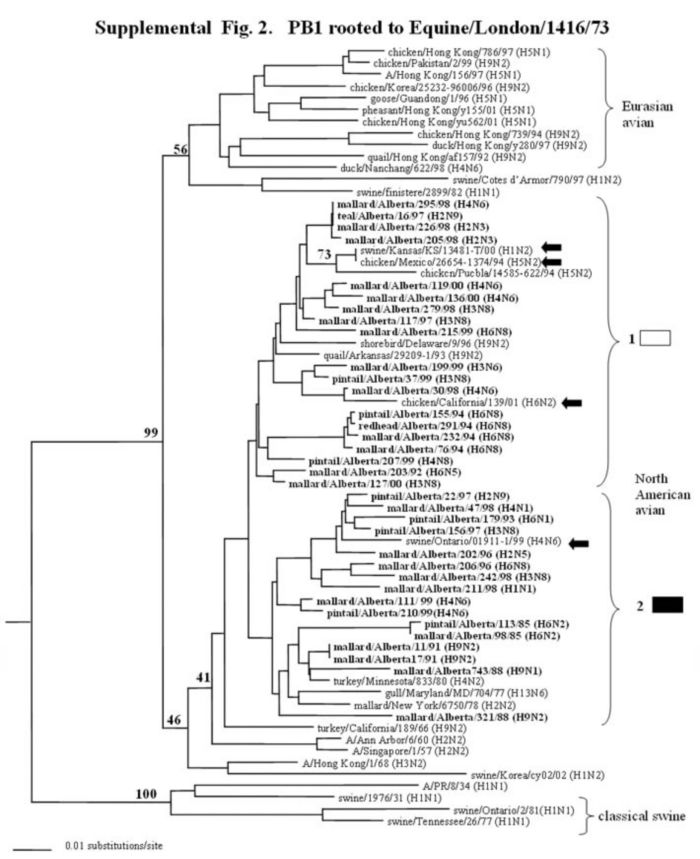
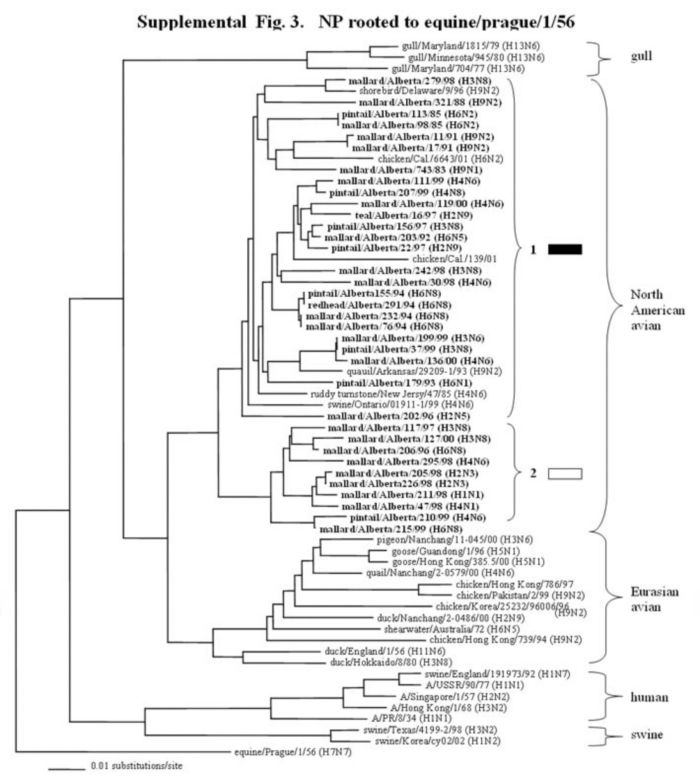
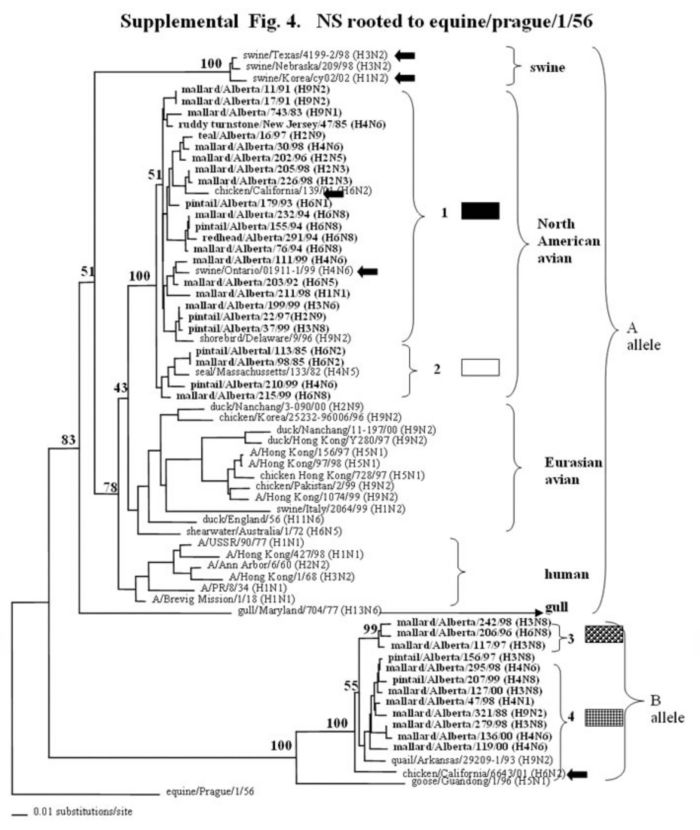
Supplemental Figure 1-5.
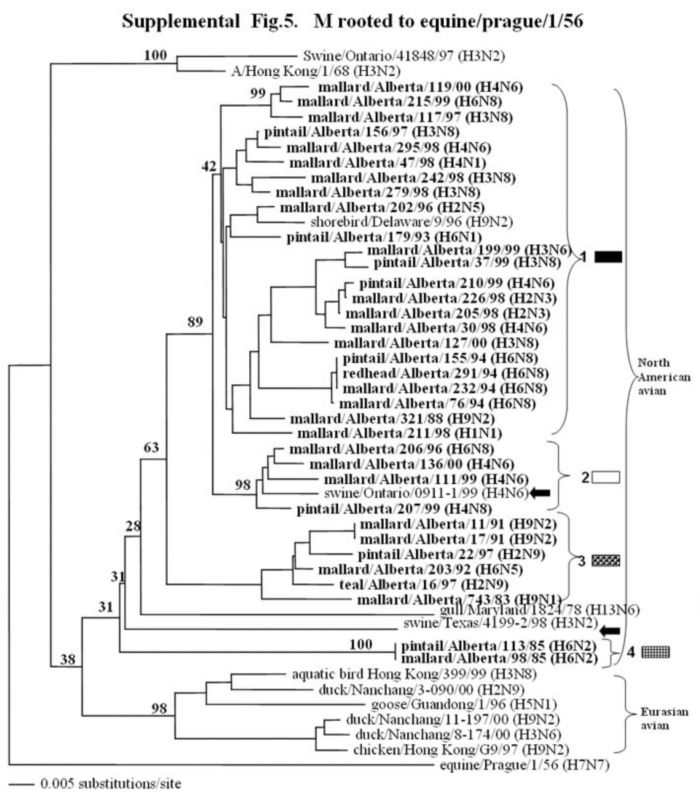
The phylogenetic trees of the internal genes (PB2 nucleotide positions 65-603; PB1, 354-565; NP, 41-631; M, 40-668; and NS, 101-621) analyzed with PAUP using bootstrap and the neighbor-joining methods as distance measures. The trees are rooted as indicated in the title. The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. The Canadian feral duck isolates sequenced in this study cluster in the North American avian clade and are in bold print; each of the distinct phylogenetic groupings is assigned a different pattern as indicated to the right; Black arrows identify swine and quail species that clustered with Canadian ducks. Abbreviations: Cal, California.
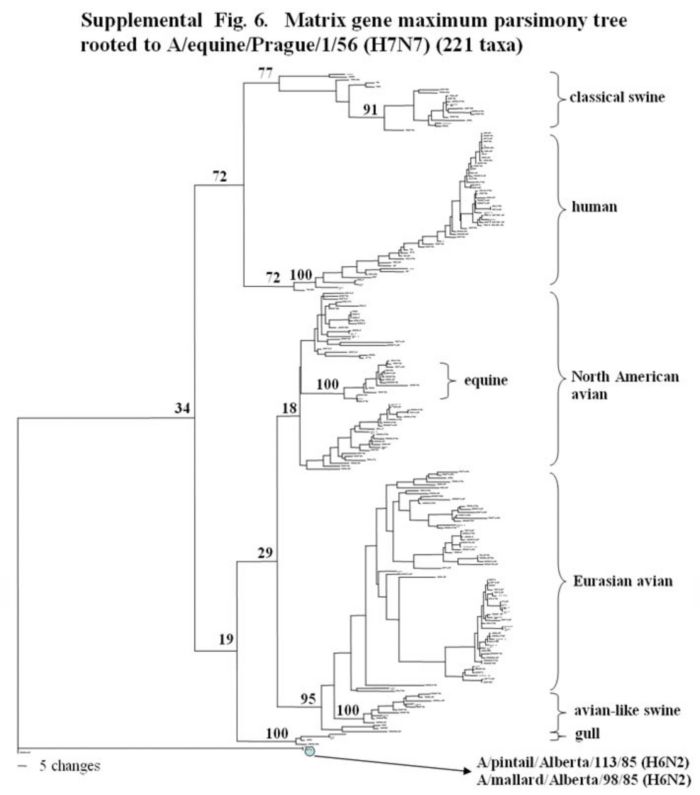
Supplemental Fig. 6.
The phylogenetic tree of the matrix gene obtained by using the complete sequences of 221 M genes (nucleotides bases 26-1004) analyzed with PAUP using the bootstrap and the maximum parsimony methods as distance measures. The tree is rooted to A/equine/Prague/56 (H7N7). The bar indicating the number of nucleotide changes is shown under the tree and bootstrap values (100 replicates) are given for selected nodes. Major phylogenetic divisions are indicated to the right of the tree and the two Canadian feral duck outliers (A/pintail/Alberta/113/85 and A/mallard/Alberta/98/85) are identified at the base of the tree.
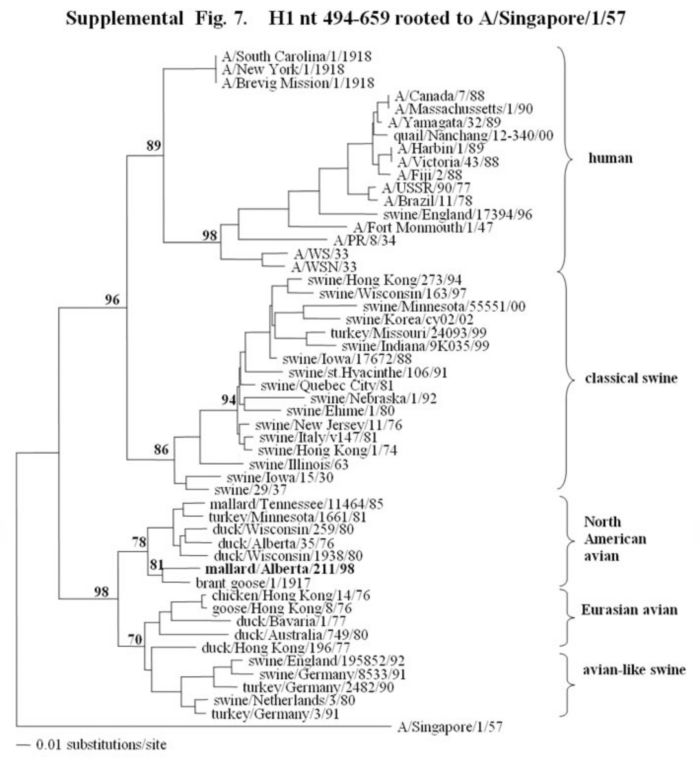
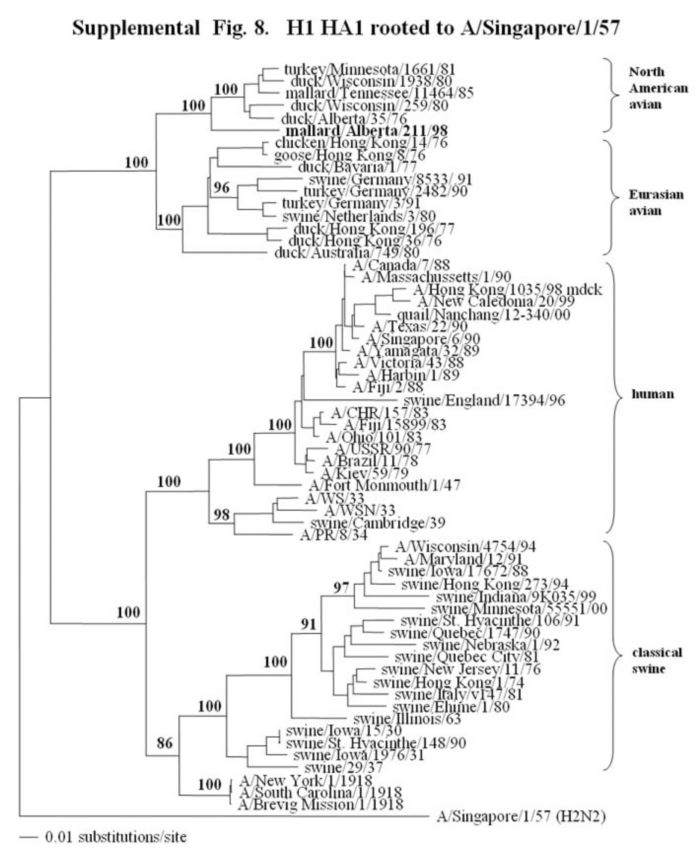
Supplemental Fig. 7,8.
The phylogenetic trees of a fragment of the H1 hemagglutinin gene (fig.7) (nucleotides bases 494-659) and the complete HA1 of the H1 hemagglutinin gene (fig.8) (nucleotides bases 33-1061) analyzed with PAUP using the bootstrap and the neighbor-joining methods as distance measures. The tree is rooted to A/Singapore/1/57 (H2N2). The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. Major phylogenetic divisions are indicated to the right of the tree. The Canadian feral duck isolate is in bold at the base of the North American avian clade.
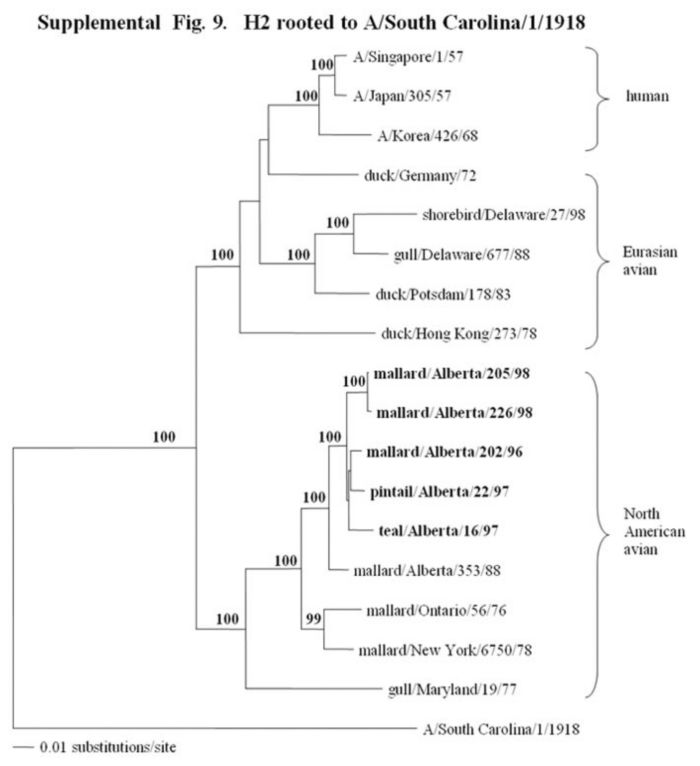
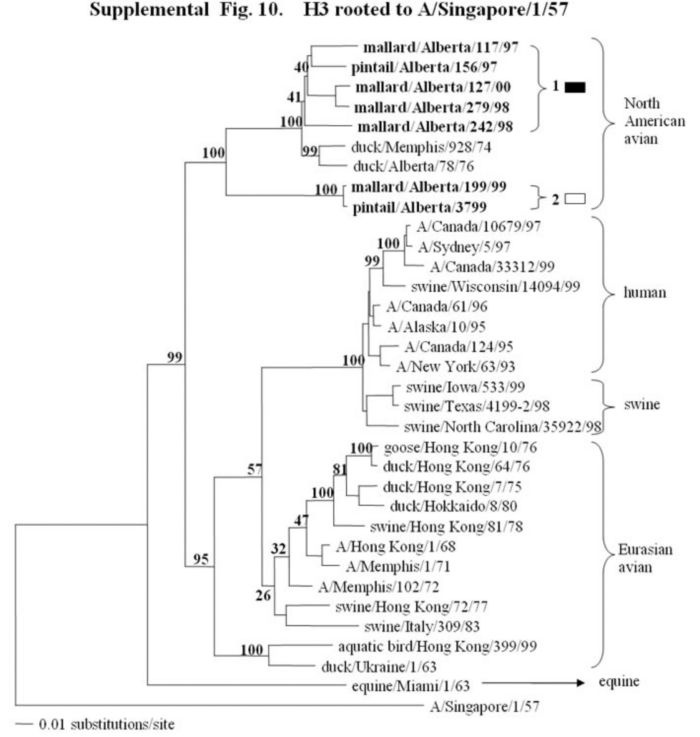
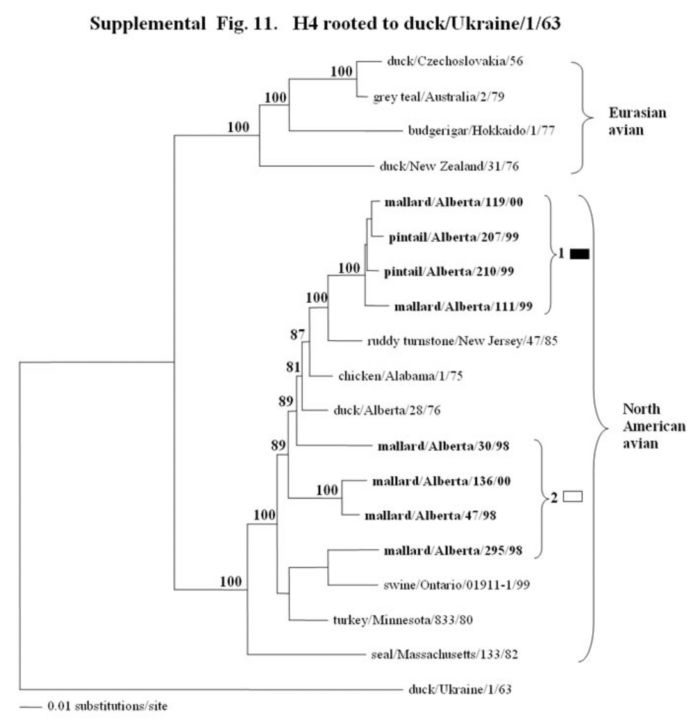
Supplemental Fig. 9-11.
The phylogenetic trees of the hemagglutinin H2, H3, and H4 genes (H2 nucleotide positions 44-1060; H3, 78-1035; and H4, 30-1647) analyzed with PAUP using bootstrap and the neighbor-joining methods as distance measures. The trees are rooted as indicated in the title. The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. The Canadian feral duck isolates sequenced in this study cluster in the North American avian clade and are in bold print. The Canadian feral duck isolates formed one phylogenetic grouping in the H2 tree and 2 separate phylogenetic groupings in the H3 and H4 trees. Each distinct phylogenetic grouping is assigned a different pattern as indicated to the right.
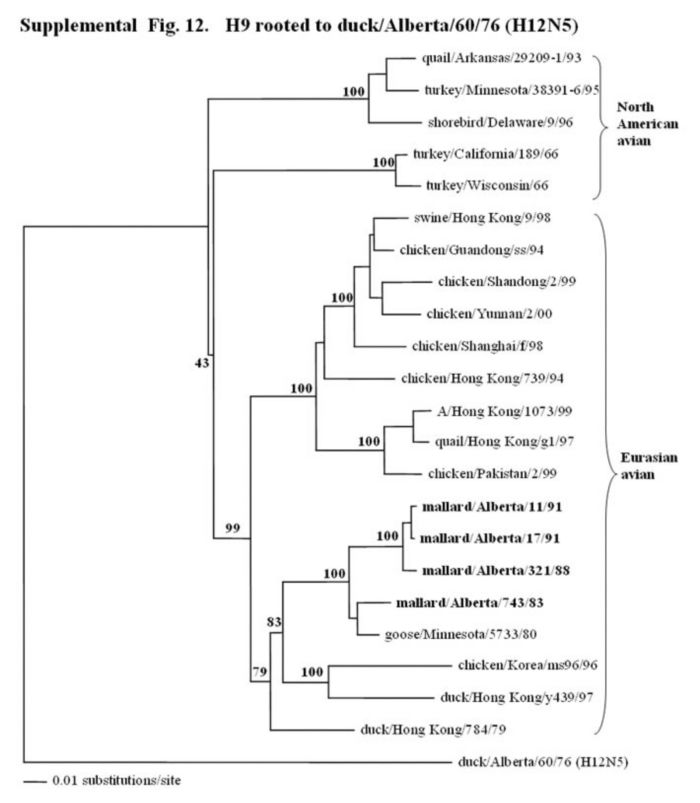
Supplemental Fig. 12.
The phylogenetic tree of the HA1 of the H9 hemagglutinin gene (nucleotides bases, 43-1047) analyzed with PAUP using the bootstrap and the neighbor-joining methods as distance measures. The tree is rooted to A/duck/Alberta/60/76 (H12N5). The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. Major phylogenetic divisions are indicated to the right of the tree. The Canadian feral duck isolates all cluster together in the Eurasian avian clade and are in bold.
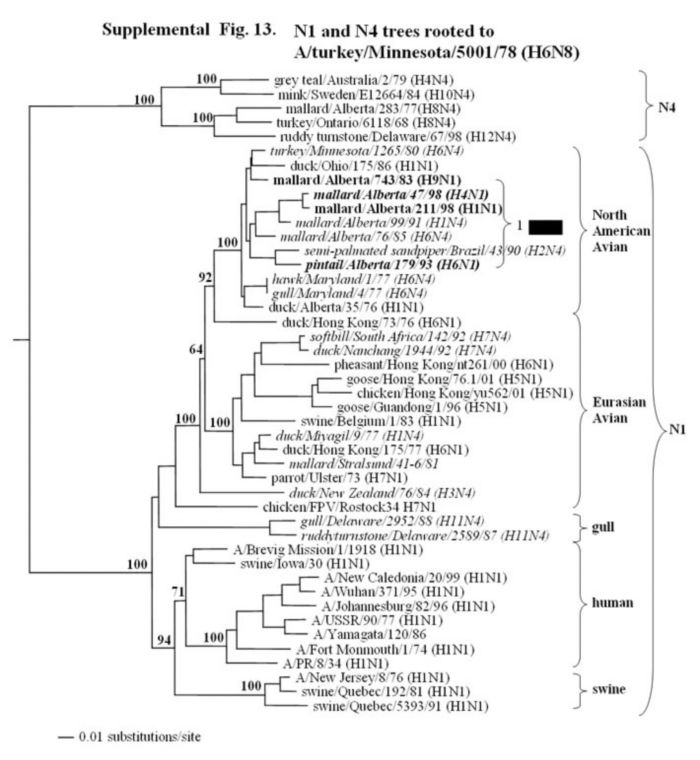
Supplemental Fig. 13.
The phylogenetic tree of the N1 gene (nucleotides bases, 21-1430) analyzed with PAUP using the bootstrap and the neighbor-joining methods as distance measures. The tree is rooted to A/turkey/Minnesota/5001/78 (H6N8). The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. Major phylogenetic divisions are indicated to the right of the tree. The Canadian feral duck isolates all cluster together in the North American avian clade and are in bold. Viruses in italics represent isolates that antigenically identified as N4 by the NA inhibition assay using reference sera to H4; however nucleotide sequence and phylogenetic analysis identified these NA genes as N1 subtypes. Included in this tree are sequences provided by Webby et al. (personal communication).
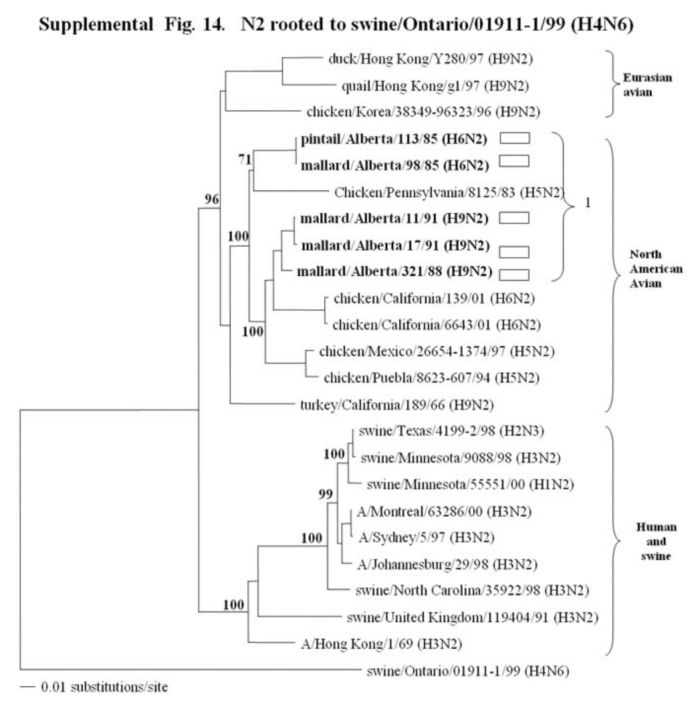
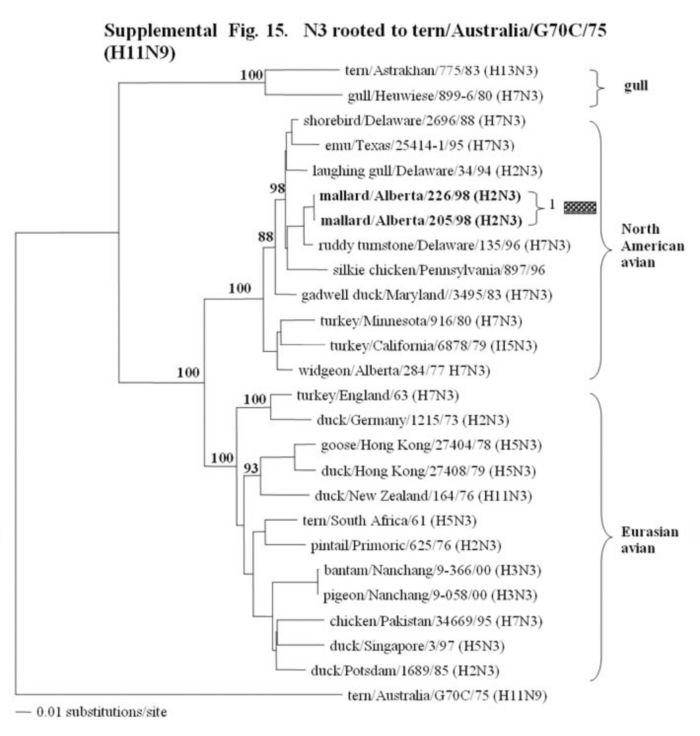
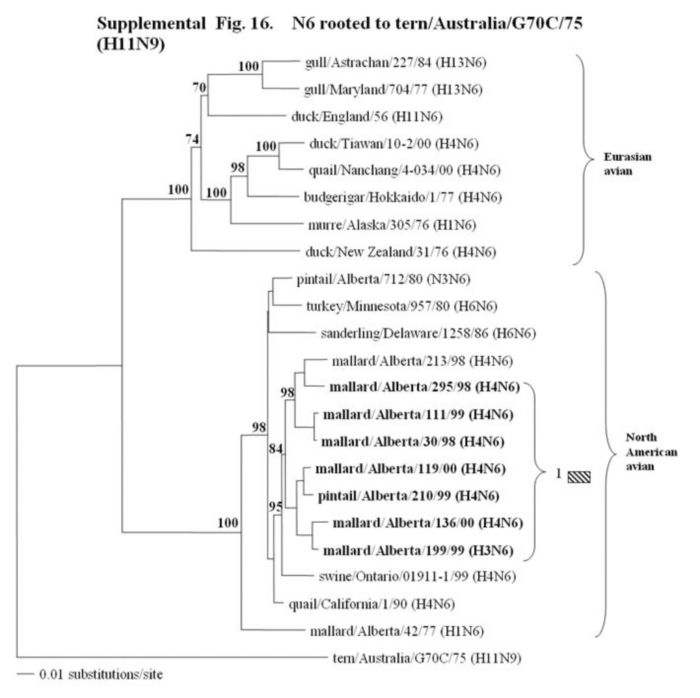
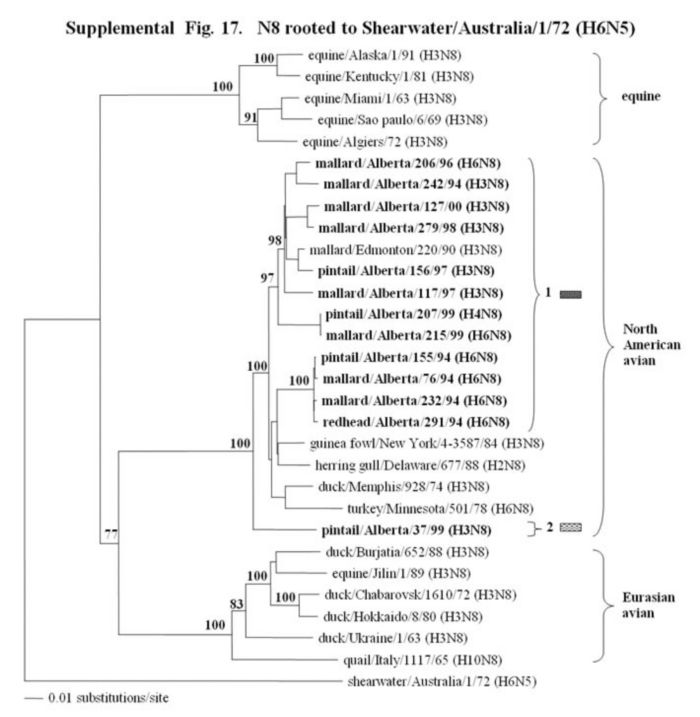
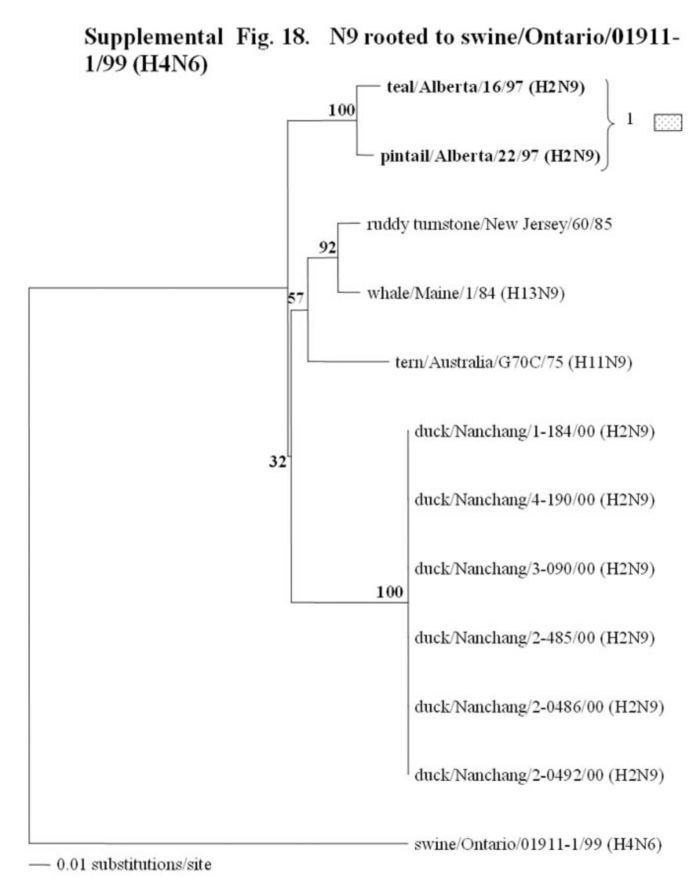
Supplemental Fig. 14-18.
The phylogenetic trees of the N2, N3, N6, N8 and N9 genes (N2 nucleotide positions 27-1395; N3, 21 1430; and N6, 21-1433; N8, 21-1415; N9, 158-601) analyzed with PAUP using bootstrap and the neighbor-joining methods as distance measures. The trees are rooted as indicated in the title. The distance bar is shown under the tree, and bootstrap values (100 replicates) are given for selected nodes. The Canadian feral duck sequenced in this study cluster in the North American avian clade and are in bold print. The Canadian feral duck N2, N3, N6 and N9 genes all clustered into one phylogenetic grouping. The N8 genes formed two distinct phylogenetic groupings. Each of the distinct phylogenetic groupings is assigned a different pattern as indicated to the right each of the viruses.