St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
The rate of growth of biological and medical data will continue to accelerate during the coming decade. Effectively managing, integrating, and exploring these data to generate new knowledge requires specialized expertise and innovative technology. St. Jude freely shares the data generated, and visualization tools engineered, with the global scientific community.
In the news
Childhood cancer genetics: A legacy of groundbreaking work drives St. Jude forward
St. Jude Children’s Research Hospital has a rich history of research on genetics stemming from the Pediatric Cancer Genome Project and reverberating through laboratories today.
Visualization and analysis tools break down barriers for big data
St. Jude is at the forefront of creating tools to visualize and analyze genomic data.
Pediatric Cancer Genome Project data
The St. Jude-Washington University Pediatric Cancer Genome Project is the world’s most ambitious effort to discover the origins of childhood cancer and seek new cures. To date, we have compared whole genomes from cancerous and normal cells for more than 800 patients, successfully pinpointing the genetic and epigenetic factors behind some of the toughest pediatric cancers.
Raw sequence data for all published results, as well as data analysis and visualization tools, are freely available. Explore them today to advance your research and help us find cures.

St. Jude Cloud
St. Jude Cloud provides data and analysis resources to the global research community. Our goal is to empower researchers across the world to advance cures for pediatric cancer and other pediatric catastrophic diseases. Partnering with Microsoft and DNAnexus, our apps are a cohesive blend of comprehensive data, scientific expertise, engineering innovation, and cloud infrastructure.

Resources from the Childhood Solid Tumor Network
The Childhood Solid Tumor Network (CSTN) offers comprehensive scientific resources and data for researchers studying pediatric solid tumors and related biology. By freely sharing unique tools with no obligation to collaborate, we aim to promote fundamental research and new treatments for pediatric patients with poorly understood and difficult-to-treat solid tumors. Resources include extensive genomic and drug sensitivity data sets, orthotopic xenografts and preclinical models.
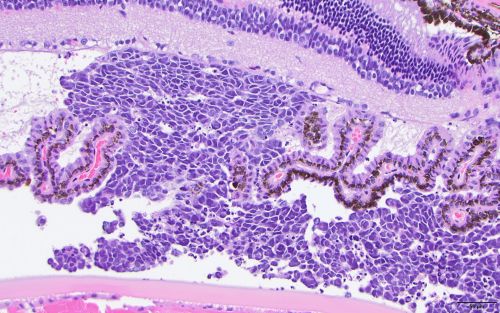
Find leukemia xenografts and data
The Public Resource of Patient-derived and Expanded Leukemias (PROPEL) offers one of the world’s largest collections of patient-derived xenografts (PDX) for adult and pediatric leukemias. Visit the PROPEL data portal to browse hundreds of available leukemia xenograft samples, submit a request and view associated genetic data for xenografts and primary tumors. Academic researchers may request PDX samples free of charge under a material transfer agreement and with no obligation to collaborate.
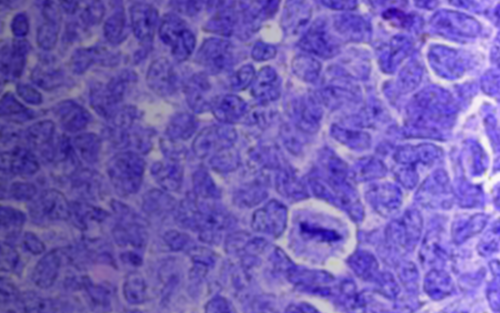
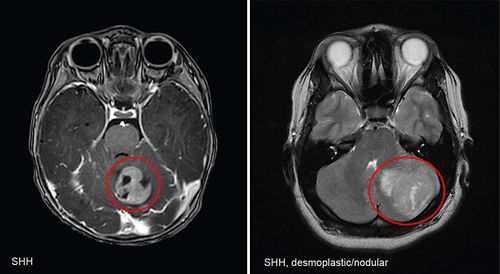
Study models of pediatric brain tumors
The Pediatric Brain Tumor Portal features molecular characterization for patient-derived orthotopic xenograft (PDOX) models of pediatric CNS tumors and reflects a nearly decade-long effort to generate and extensively characterize in vivo models that faithfully recapitulate pediatric brain cancers. The portal offers visualization tools that allow users to interrogate curated datasets and access models from our PDOX library for functional studies of tumorigenesis or preclinical testing.
Research computing
Research Information Services (RIS) at St. Jude Children's Research Hospital provides high-performance scientific computing systems, computational resources, scientific software applications and computational and software engineering services to support the St. Jude research community.




