St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Available Resources
The Childhood Solid Tumor Network (CSTN) at St. Jude offers a comprehensive collection of scientific resources for academic researchers studying pediatric solid tumors and related biology. An overview of available samples and data is provided below; visit the CSTN data portal to request samples and for more detailed information. To receive updates on new resources emerging from the CSTN, subscribe to our e-newsletter.
Available resources include:
Submit a request to receive research data and samples.
More information
If interested in GEMMs, please send an email to CSTN@stjude.org.
Protocols with various antibodies, targets and tissue types have been developed and validated at St. Jude. Access resources.
Orthotopic patient-derived xenografts (O-PDXs) have been created from a wide variety of pediatric solid tumors collected under the Molecular Analysis of Solid Tumors (MAST) protocol at St. Jude Children’s Research Hospital (ClinicalTrials.gov: NCT01050296). Most O-PDX models are available with a luciferase reporter gene integrated into the genome to enable monitoring of tumor engraftment and growth in vivo. Cryopreserved cells, fresh frozen tissue/cells, and formalin-fixed paraffin-embedded tissue blocks are available.
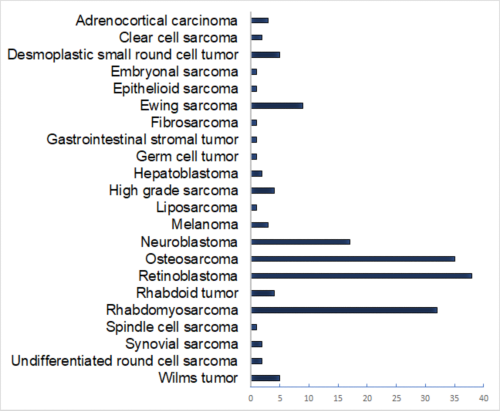
Access CSTN Data Portal for the full list of tumor types and samples. Extensive molecular and drug sensitivity data are available for the O-PDX models. For each of the O-PDX models, a characterization sheet will be provided when the samples are shipped (see example for SJNBL046_X or visual descriptions of the clonal evolution groups.) Characterization sheets can also be downloaded for each sample in the CSTN Data Portal to help investigators select the appropriate O-PDX models for their studies. For the samples that have been added most recently, characterization is pending and the completed characterization sheets will be sent to requestors as soon as they are available.
The list of available tumors is updated every year after the new samples have engrafted and sufficient vials have been cryopreserved for distribution.
Induced pluripotent stem cells (iPSCs) have been created from 15 patients with retinoblastoma that have germline RB1 mutations collected under the RETCELL (A Study of Children with Heritable Retinoblastoma) protocol at St Jude Children’s Research Hospital (ClinicalTrials.gov NCT02193724). The clinical spectrum of disease varies from early diagnosis (Group A and/or B) to advanced bilateral disease (Group D and/or E). In addition, some participants were found to be unaffected or spontaneously regressed carriers of the heritable mutation or chromosomal abnormality. The RB1 mutations have been validated in each line and they have undergone complete characterization including trilineage differentiation, whole genome sequencing, karyotyping and 3D retinal organoid differentiation. Vials of cryopreserved cells are available upon request. Additional iPSC lines are being developed for other pediatric solid tumor types and the list of available lines will be updated every six months after they have been validated and characterized.
To find out more, email CSTN@stjude.org.
Genomic, drug sensitivity and pharmacokinetics data
We offer molecular and drug sensitivity data of diverse pediatric solid tumors, patient-derived orthotopic xenografts and solid tumor cell lines. (Some cell lines are available. Visit the CSTN data portal for details) High-throughput drug screening (HTS) data are available for many pediatric solid tumor cell lines and orthotopic xenografts, including Ewing sarcoma, neuroblastoma, osteosarcoma, retinoblastoma and rhabdomyosarcoma.
While comprehensive raw datasets are not made available prior to publication, we are happy to accommodate many types of data requests to answer specific research questions.
Raw datasets are available through St. Jude Cloud, following an application and approval process for access.
Genomic sequencing and integrated epigenetic profiling have been performed on pediatric solid tumors and orthotopic patient-derived xenografts as part of the St. Jude-Washington University Pediatric Cancer Genome Project and is actively ongoing in St. Jude investigator-led labs. Available data may include:
- Whole-genome sequencing (WGS)
- Whole-exome sequencing (WES)
- RNA-seq
- Whole-genome bisulfite sequencing (WGBS)
- ChIP-seq
High-throughput drug screening (HTS) data are available for many pediatric solid tumor cell lines and orthotopic xenografts, including Ewing sarcoma, neuroblastoma, osteosarcoma, retinoblastoma and rhabdomyosarcoma. In addition, we offer preclinical pharmacokinetic data for a number of high-priority drugs that are being explored for activity against pediatric solid tumors. Standard of Care regimens used in current preclinical trials are available in the appendix. Data from completed preclinical Phase I (tolerability), Phase II (small pilot efficacy study) and Phase III (randomized, placebo-controlled) trials are available by request. For more details and descriptions of Phase I, II and III, please see Stewart et al, 2014 Cell Reports.
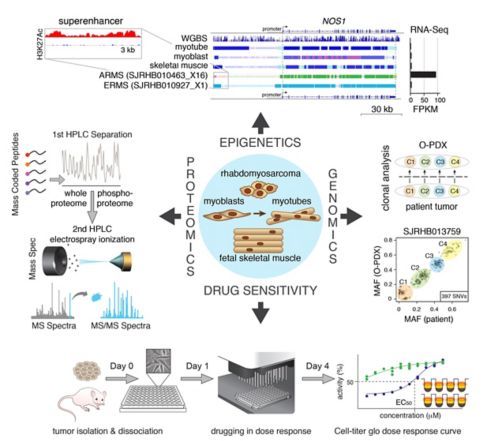
We have collected and integrated genomic, epigenetic, proteomic and drug-sensitivity data into a central database. The genomic data includes whole genome, exome and RNA-sequence, as well as clonal analysis that allows us to reconstruct the clonal heterogeneity of the tumor and corresponding O-PDX. The epigenetic data includes whole genome bisulfite (WGBS) and ChIP-seq data for eight histone marks and CTCF, Brd4 and RNA PolII for a total of over 750 ChIP-seq libraries. The data have been integrated to define chromatin states using chromatin Hidden Markov Modeling. Quantitative proteomic and phosphoproteomic data were obtained for RMS tumors using mass coded peptides derived from batches of 10 samples. Drug-sensitivity data includes primary cultures of O-PDX tumors in high throughput drug screening of 168 oncology drugs in 10-point dose response in triplicate with biological replicates.
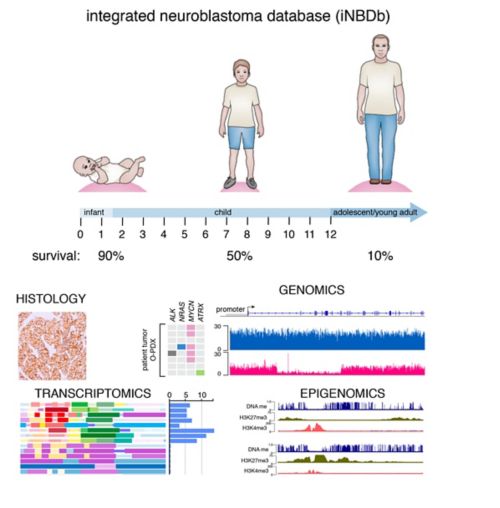
We have collected and integrated genomic, epigenetic, proteomic and drug-sensitivity data into a central database for neuroblastoma. We have focused on age at diagnosis because there is an inverse relationship between age at diagnosis and outcome. Moreover, ATRX mutations are significantly associated with older age at diagnosis so we have included several ATRX mutant NBs. The genomic data includes whole genome, exome and RNA-sequence (transcriptome) as well as clonal analysis that allows us to reconstruct the clonal heterogeneity of the tumor and corresponding O-PDX. The epigenetic data includes whole genome bisulfite (WGBS) and ChIP-seq data for eight histone marks and CTCF, Brd4 and RNA PolII for a total of over 350 ChIP-seq libraries. The data have been integrated to define chromatin states using chromatin Hidden Markov Modeling. Drug-sensitivity data includes primary cultures of O-PDX tumors in high throughput drug screening of 168 oncology drugs in 10-point dose response in triplicate with biological replicates.
About the Childhood Solid Tumor Network
Childhood solid tumors are often difficult to study and treat because they are rare and originate in the complex biological context of developing organs. CSTN was established to disseminate resources and data that have been developed at St. Jude Children’s Research Hospital, with the aim of stimulating basic research and speeding translation to the clinic.
The effort was launched in 2013 by Howard Hughes Medical Institute investigator Michael Dyer, PhD, of St. Jude Developmental Neurobiology, and Alberto Pappo, MD, of St. Jude Oncology.
For more information, contact us: CSTN@stjude.org
Connect with CSTN

Receive updates on emerging resources and research advances by subscribing to our e-newsletter.